
Artigo original
HOLOCENTRIC CHROMOSOMES IN ANIMALS AND PLANTS: WHAT DO WE KNOW ABOUT THESE POINTS OUTSIDE THE CURVE?
Cromossomos holocêntricos em animais e plantas: o que nós sabemos sobre estes pontos fora da curva?
Gabriela Corrêa Morais*
 Thainara Siqueira Messias†
Thainara Siqueira Messias† Giancarlo Conde Xavier Oliveira‡
Giancarlo Conde Xavier Oliveira‡
https://doi.org/10.18593/evid.32760
Recebido em 14 de maio de 2023 | Aceito em 31 de agosto de 2023
Abstract:
Usually, members of the Eukarya domain have kinetochores grouped at a single point, characterizing monocentric chromosomes. However, in some taxa the protein apparatus that composes the kinetochore is distributed continuously or discretely along the length of the chromosome, condition that defines holocentric chromosomes. The review aims to provide an overview of karyomorphological aspects of holocentric chromosomes in animals and plants, as well as an understanding of their origin, evolution and possible adaptive implications of their presence in eukaryotes. The main structural differences between holocentric and monocentric chromosomes concern the kinetochore proteins, the pattern of histone H3 phosphorylation and centromeric satellite DNA. The distribution of holocentric chromosomes in phylogenetic trees evidence their independent emergence numerous times throughout evolution. While many hypotheses have been created to explain the origin of holocentric chromosomes, none have been confirmed or refuted. Although the adaptive advantages generated by their presence are undeniable, especially in clastogenic environments, the typical behavior of chromosomes with diffuse kinetochores seems to be enough to make them the exception rather than the rule among eukaryotes.
Keywords: Centromere. Kinetochores. CENH3. Luzula nivea. Caenorhabditis elegans.
Resumo:
Usualmente, membros do domínio Eukarya possuem cinetócoros agrupados em um único ponto, o que caracteriza os cromossomos monocêntricos. Todavia, em alguns taxa o aparato proteico que compõe o cinetócoro é distribuído continuamente ou discretamente ao longo do comprimento do cromossomo, condição que define os cromossomos holocêntricos. O objetivo desta revisão é fornecer uma visão geral dos aspectos cariomorfológicos dos cromossomos holocêntricos de animais e plantas, assim como uma compreensão de sua origem, evolução e possíveis implicações adaptativas da sua presença em eucariotos. As diferenças estruturais mais consideráveis entre cromossomos holocêntricos e monocêntricos dizem respeito às proteínas do cinetócoro, ao padrão de fosforilação da histona H3 e ao DNA satélite centromérico. A distribuição de cromossomos holocêntricos em árvores filogenéticas evidencia seu surgimento independente inúmeras vezes ao longo da evolução. Apesar de muitas hipóteses terem sido criadas para explicar a origem dos cromossomos holocêntricos, nenhuma foi confirmada ou refutada. Embora sejam inegáveis as vantagens adaptativas geradas por sua presença, especialmente em ambientes clastogênicos, o comportamento típico dos cromossomos com cinetócoros difusos parece ser suficiente para torná-los a exceção e não a regra entre os eucariotos.
Palavras-chave: Centrômero. Cinetócoros. CENH3. Luzula nivea. Caenorhabditis elegans.
@Autor correspondente: Master in Genetics and Plant Breeding. “Luiz de Queiroz” College of Agriculture, University of São Paulo, Genetics Department, Pádua Dias Avenue, 11, CEP 13400-970, Piracicaba, São Paulo, Brazil; https://orcid.org/0000-0002-9532-9057; gabrielacm@usp.br.
1 INTRODUCTION
Centromeres, first described by the cytologist Whalter Flemming in 1882, plays a key role in the separation of chromosomes during cell division. When active, they have a highly specialized disk-shaped protein arrangement called kinetochore, which interacts with tubulins, constituent elements of spindle fibers. Once anchored to the kinetochores, microtubules ensure the movement of chromatids and chromosomes to the poles during mitotic and meiotic events1. Centromeres, especially in the kinetochore region, form a constriction visible under microscopy in metaphase chromosomes called primary constriction, considered an important chromosomal marker2.
Usually, members of the Eukarya domain have kinetochores mounted at a single point, characterizing monocentric chromosomes. However, at least two centromere morphologies exist in eukaryotic organisms. In some taxa, the protein apparatus that composes the kinetochore is distributed continuously or discretely along the length of the chromosome, allowing the spindle fibers to connect along all or almost all of its longitudinal extent3-4. This condition defines holocentric chromosomes (term adopted in this literature review), also called chromosomes with diffuse kinetochores, polykinetic chromosomes, holokinetic chromosomes, dicentric chromosomes or semilocalized chromosomes5.
First characterized in 1935 by Franz Schrader and his wife Sally Hughes-Schrader in members of the order Hemiptera6, holocentric chromosomes were previously described as chromosomes connected by a delicate filame, with some “confused” stages during meiosis7. Although reported in a minority of eukaryotes (including animals, plants, fungi and algae), their prevalence may be underestimated, given the large number of species not yet cataloged and the difficulty in studying some life forms cytologically. Historically, the most commonly used strategy for its identification is the adoption of cytological methods that allow the observation of the behavior of chromosomes during mitosis or meiosis. Chromosomal fragmentation techniques using gamma rays or X-rays and subsequent microscopic observation of the dividing cells are also usually applied, although, in both cases, they are only viable when we are dealing with species with large and few chromosomes. Molecular techniques – such as those that allow the identification of kinetochore proteins – have been used in the case of species with short and numerous chromosomes8. Measuring the nuclear DNA content of meristems exposed to gamma radiation using flow cytometry also constitutes a viable non-microscopic strategy for distinguishing between monocentrism and holocentrism9, but it is not a usual tool for all organisms, as it is not yet widely implemented.
In addition to the position of the kinetochore, holocentric chromosomes manifest other peculiarities. Unlike monocentric chromosomes, they are marked by the absence of visible primary constriction and do not have pericentromeric regions rich in heterochromatin – whose quantity and distribution are extremely variable10. They also have homogeneous condensation, although this is not an exclusive characteristic of the group11. Furthermore, they exhibit intriguing behaviors during mitosis and meiosis regarding their positioning along the equatorial plate and migration to the cell’s poles.
This review aims to provide an overview of karyomorphological aspects of holocentric chromosomes in animals and plants, as well as an understanding of their origin, evolution and possible adaptive implications of their presence in eukaryotic organisms.
2 MORPHOLOGY AND BEHAVIOR OF HOLOCENTRIC CHROMOSOMES
Spindle fibers attached to a functional kinetochore that extends along its entire length or nearly so during cell division constitute the main feature of holocentric chromosomes. Studies on the distribution of heterochromatin in species of the genus Luzula known to be holocentric have evidenced the existence of C bands along their entire length12. However, subsequent investigations showed that in other holocentric taxa there is a higher concentration of constitutive heterochromatin in terminal regions and a lower concentration in interstitial and median regions13. Thus, there seems to be no constitutive heterochromatin pattern that allows the discrimination of holocentric chromosomes. Although ribosomal DNA (rDNA) sites or the Nucleolus Organizer Region (NOR) in holocentric chromosomes usually occupy terminal regions, they can also be found in interstitial regions as well as monocentric chromosomes, where they are more often subterminal or interstitial14-15. Both conditions suggest that the distribution of the centromeric region presumably has little or no influence on chromatin organization (and vice versa).
In fact, the main structural differences between holocentric and monocentric chromosomes concern the kinetochore proteins, the pattern of histone H3 phosphorylation and centromeric satellite DNA. Kinetochore proteins are extremely conserved in eukaryotes, especially CENH3 (a generic term used to name a histone H3 variant), which plays an important role in this apparatus. Using an immunostaining technique with anti-CENH3 antibodies in metaphase chromosomes of Luzula nivea, it was possible to identify that this key protein externally covers a large part of the length of each sister chromatid in holocentric chromosomes, with the exception of telomeric regions. Furthermore, the amount and distribution of CENH3 in holocentric chromosomes vary throughout the different phases of the cell cycle, which does not occur in monocentric chromosomes. In interphase nuclei containing chromosomes with diffuse centromeres, CENH3 is found in a dispersed manner and at specific points, increasing considerably during prophase and metaphase and decreasing gradually during anaphase, telophase and interphase16-17. Histone H3 phosphorylation is crucial for maintaining cohesion between sister chromatids during the cell cycle. Although it is a highly conserved event, it can manifest some particularities in different groups of organisms18. In monocentric plant species, histone H3 is phosphorylated along the entire chromosome during prophase I of the first meiotic division, which causes sister chromatids to hold together along its entire length. During mitosis and second meiotic division, phosphorylation is limited to sites close to the centromeric region19. In contrast, in holocentric chromosomes of species such as Luzula luzuloides and Rhynchospora tenius, both in mitosis and in the first and second meiotic divisions, histone H3 phosphorylation occurs homogeneously throughout its length, ensuring strong cohesion between the chromatids during these cycles20-21.
In eukaryotes, centromeric satellite DNA is characterized by being highly variable in terms of size, number of repeats, as well as the sequence of nucleotides that compose them. Investigations involving holocentric species of the genus Luzula have shown that centromeric satellite DNA is distributed in the form of multiple sites along chromosomes and that the size of its repeat units varies even within species – for example, 175 to 178 bp in Luzula nivea22. In monocentric chromosomes, the number of base pairs that form the repeat units of this satellite DNA appears to be fixed and species-specific23-24.
In addition to structure, the presence of diffuse centromeres also has implications for the behavior of chromosomes throughout the cell cycle. Its movement during the prometaphase of mitosis in C. elegans is faster due to efficient capture of microtubules25. Furthermore, there is an imbalance in the forces applied under the sister kinetochores, which does not compromise, however, the coordinated work between them. The more pronounced tension or pulling force under holocentric chromosomes ultimately requires greater chromatin compaction during prophase, resulting in stiffer chromatids. This rigidity, combined with a strong longitudinal cohesion of chromatids, prevents the same kinetochore from being connected to microtubules at both poles of the spindle, thus reducing the chances of aneuploidies. During anaphase, sister chromatids migrate to opposite poles along all or most of their length without loss of orientation parallel to the equatorial plate, which deviates from the “V” conformation characteristic of monocentric chromosomes (Figure 1)26-27.
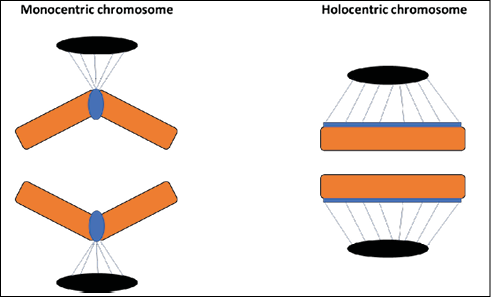
Figure 1 – Conformation of sister chromatids of monocentric and holocentric chromosomes during anaphase of mitosis. On the left: monocentric chromosome with sister chromatids in a “V” conformation and spindle fibers anchored to a localized centromere. On the right: holocentric chromosome with sister chromatids in parallel conformation and spindle fibers anchored along its entire length.
As mentioned earlier, in both monocentric and holocentric chromosomes, sister chromatids are held together along their entire length during prophase I of the first meiotic division. However, unlike holocentric chromosomes, during the subsequent stages of meiosis I, the cohesion between chromatids in monocentric chromosomes is lost, remaining only near the regions corresponding to centromeres and chiasmata28. These remains of cohesion last for a large part of the cycle, being eliminated only during anaphase II of the second meiotic division.
In holocentric chromosomes, only one or two chiasmata are found per bivalent, regardless of the size of the chromosomes. Furthermore, recombinations tend to occur in more distal regions, poor in terms of the number of genes29-30. By analyzing the meiotic behavior of holocentric chromosomes of Psylla foersteri Flor. (Psylloidea, Hemiptera), it was possible to verify normal meiosis in bivalents with one or two chiasmata, whose occurrence was more frequent. Bivalents with three chiasmata were unable to complete anaphase I due to non-resolution of the internal or central chiasma, resulting in abnormal meiosis. This scenario ends up creating a selection pressure against the formation of multiple chiasmata – which seem to face a structural barrier apparently inexistent in monocentric chromosomes – and explains the higher frequency of a single recombination by bivalent characteristic of holocentrism31.
Meiosis in holocentric chromosomes also carries particularities regarding how the microtubules of the spindle fibers are attached to the sister chromatids, which will affect their movement pattern during anaphase II. If during meiosis I the bivalent is positioned parallel to the equatorial plate (equatorial orientation), the chromatids are pulled to opposite poles along their entire length in anaphase II, as in the mitotic cycle of all chromosomes with diffuse kinetochores32. This behavior defines holokinetic meiosis, a typical condition of the meiotic cycle of holocentric plant species, including Luzula campestris33. However, if the bivalent has an axial orientation (positioned perpendicularly to the equatorial plate), the microtubules are anchored at the ends of each pair of chromatids, which then play a role similar to a localized centromere, being pulled from them. This kinetic activity defines telokinetic meiosis, most frequently described in holocentric animal species, as in Caenorhabditis elegans (Figure 2)34-35.
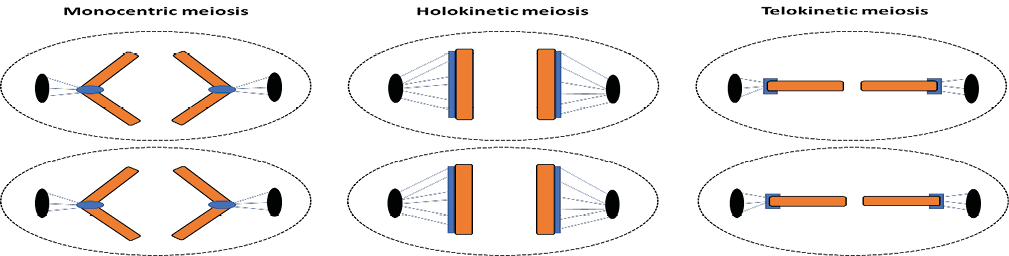
Figure 2 – Schema of the behavior of sister chromatids during anaphase II. On the left: anaphase II in monocentric chromosomes, with sister chromatids assuming a “V” conformation. In the center: anaphase II in holocentric chromosomes, in which the orientation of the bivalent along the metaphase plate is equatorial, characterizing the holokinetic meiosis. On the right: anaphase II in holocentric chromosomes, in which the orientation of the bivalent along the metaphase plate is axial, characterizing the telokinetic meiosis.
3 HOLOCENTRIC CHROMOSOMES IN ANIMALS AND PLANTS
Holocentric chromosomes have been described in about 768 species, including 540 animals and 228 plants. In the kingdom Animalia, its presence has been detected only in members of the phyla Arthropoda (including myriapods, arachnids and insects) and Nematoda8. Among the arthropods, species representing the superclass Myriapoda stand out – including the millipedes Thereuonema hilgendorfi and Thereuopoda clunifera and the centipede Esastigmatobius longitarsis Verhoe36. In the subclass Arachnida, chromosomes with diffuse kinetochores have been described in spiders (including the species Dysdera longirostris37, Segestria florentina and Dysdera crocata)38, scorpions (including Tytius bahiensis, Tytius maranhensis, Tytius mattogrossensis and Tytius paraguayensis)39-40, as well as ticks (including Rhipicephalus microplus or cattle tick) and mites (including Tetranychus urticae or two-spotted spider mite)41-42.
Among the class Insecta (taxon with the largest number of species in the animal kingdom), chromosomes with diffuse kinetochores are relatively well spread, having been characterized for the first time by Franz Schrader and Sally Hughes-Schrader in members of the order Hemiptera6. Its presence has also been observed in representatives of the Odonata orders (including the dragonflies Somatochlora metallica, Sympetrum vulgatum and Libellula haste)43-44, Psocoptera (including the book lice Blaste conspurcata and Psococerastis gibbosa) and Lepidoptera (including the domesticated silk moth Bombix mori, the wild silk moth Bombix mondarina and the butterfly Leptidea sinapis45-47. In Nematoda, holocentric chromosomes have already been identified in species such as Trichinella spiralis and Meloydogine hapla48, in addition to Caenorhabditis elegans, the (until then) only recognized holocentric model organism. This nematode, which has approximately 20 kinetochores along each of its chromosomes49, is the most widely studied species with diffuse centromeres, allowing a better understanding of the dynamics of holocentric chromosomes in other eukaryotes.
For animal species, identification of holocentric chromosomes usually occurs through observation of mitotic or meiotic cells and the behavior of sister chromatids during cell division. However, the number and/or size of chromosomes may make the adoption of this strategy unfeasible. A higher concentration of constitutive heterochromatin in the telomeric regions evidenced by the C-banding technique is a common condition in chromosomes with diffuse kinetochores50. However, this heterochromatic distribution pattern cannot properly be considered a rule, with exceptions among holocentric species51, and cannot be adopted as a criterion to identify them. In these cases, it is preferable to adopt molecular techniques that allow the identification of kinetochore proteins (including CENH3), and their distribution pattern can be used to distinguish between diffuse centromeres and localized centromeres52.
In the kingdom Plantae, holocentric species were identified only among higher plants (angiosperms), including monocots and dicots. Among monocots, holocentric chromosomes are present in representatives of Cyperaceae (e. g., species of the genus Carex), Liliaceae (e.g., Chionographis japonica Maxim) and Juncaceae53-54. A highlight is given to Luzula nivea (Juncaceae), the most widely studied holocentric plant species until a few years ago. Currently, studies are more concentrated on the genus Rhynchospora (Cyperaceae)55-57. Initially, it was believed that holocentrism would be restricted to only two genera among eudicots: Drosera (Droseraceae) and Cuscuta (Convolvulaceae). However, the species Trithuria submersa (Hydatellaceae) and Prionium serrato (Thurniaceae) were later identified as having diffuse kinetochore chromosomes9,58. Furthermore, cytogenetic analyses suggest that the dicot Myristica fragrans Houtt. (Myristicaceae) – popularly known as nutmeg, from which the spice is obtained – does not have a localized centromere, being (until then) the only recognized holocentric species with agronomic importance59.
Recently, a study involving centromere-specific antibodies and high-resolution microscopy suggested the classification of plant holocentric centromeres into two categories based on spindle fiber attachment sites: “cluster-like holocentromere” and “line-like holocentromere”. In “cluster-like holocentromere”, the kinetochore proteins CENH3 and CENP-A are distributed in groups uniformly dispersed along the chromosome, with the spindle fibers capable of binding along its entire longitudinal length. In “line-like holocentromere”, kinetochore proteins are linearly distributed along grooves, under which microtubules are anchored. In holocentric chromosomes, spindle fibers can still bind to regions that are not equivalent to those occupied by the proteins that typically make up the kinetochore. This third pattern is observed in Cuscuta europaea60.
In addition to microscopic and molecular strategies, the distinction between holocentric and monocentric chromosomes in plants is usually performed using flow cytometry. Meristematic tissues are subjected to ionizing radiation and the number of nuclei in G1 (2C) and G2 (4C) are counted by cytometry, and the G2/G1 ratio is calculated. In monocentric plants, the induction of chromosome fragmentation leads to the interruption of the cell cycle in G2, in order to prevent cells from dividing with their structurally altered chromosomes. Thus, the G2/G1 ratio tends to vary between plants exposed and not exposed to radiation. The same does not occur (or occurs to a lesser extent) in plants with holocentric chromosomes, thanks to their characteristic fragmentation tolerance (see topic “Adaptative implications of holocentric chromosomes in animals and plants”)61. Flow cytometry also allows measuring the nuclear DNA content of cells induced by chromosomal fragmentation, being also a viable alternative for the distinction between holocentric and monocentric plants9.
4 ORIGIN AND EVOLUTION OF HOLOCENTRIC CHROMOSOMES
The distribution of holocentric chromosomes in phylogenetic trees in distant and supposedly unrelated lineages evidence their independent emergence numerous times throughout evolution, with no transitional forms between monocentrism and holocentrism. The primitive condition of the centromeres of eukaryotic organisms is still uncertain62. Some authors theorize that diffuse kinetochores constitute a plesiomorphic condition, while localized kinetochores are an apomorphic condition63. However, the fact that holocentric chromosomes in plants are restricted to angiosperms leads us to believe that, at least among plants, holocentrism is a derived character, being a homoplasy resulting from parallelism that arose independently at least four times in the kingdom Plantae64.
Many hypotheses have been created to explain the origin of holocentric chromosomes. The oldest of them suggests that holocentrism is the result of the propagation of centromeric regions to several loci from the movement of transposable elements, the occurrence of pericentric inversions and chromosomal duplications64. A second hypothesis suggests that holocentrism arose from telomeres along the evolution of linear chromosomes, originated from the breaking of an ancestral circular genome – telomeric sequences until then scattered in internal regions of the newly emerged linear chromosome began to accumulate and form subtelomeric sequences recognized by tubulin, giving rise to centromeres65. The presence of protein complexes recognized by actin that act in the segregation of chromosomes in prokaryotes (remains of the old circular genome) coexisting with the newly emerged centromeres led to instability in the first linear chromosomes, which, after successive cycles of breaks and amplifications, evolved into monocentric or holocentric states66. This “telomere to centromere” model is supported by the existence of tandem telomeric repeats in interstitial regions in genomes of holocentric species (including the nematode Caenorhabditis elegans), although they are more accumulated at the ends of chromosomes67. A third hypothesis suggests that satellite regions of monocentric chromosomes would have spread along their longitudinal extension, triggering multiple centromeres and giving rise to holocentric chromosomes. This idea is supported by the existence of centromeric satellite DNA sequences common between rice (whose kinetochore is localized) and Luzula nivea (which is known to have diffuse kinetochores)68.
5 ADAPTATIVE IMPLICATIONS OF HOLOCENTRIC CHROMOSOMES IN ANIMALS AND PLANTS
Scientists have long questioned whether monocentric and holocentric chromosomes are neutral in terms of evolutionary success or whether both may represent adaptive advantages in specific scenarios69. Although conclusive evidence is still lacking, it is known that holocentrism offers eukaryotic lineages the advantageous ability to respond better to structural chromosomal damage70. Breaks in monocentric chromosomes result in the formation of acentric fragments, which tend to be lost during cell division1. In contrast, the presence of diffuse centromeres along the entire (or almost the entire) length of holocentric chromosomes means that the fragments generated after their breakage can still be recognized by the spindle fibers, maintaining their kinetic activity (Figure 3)71.
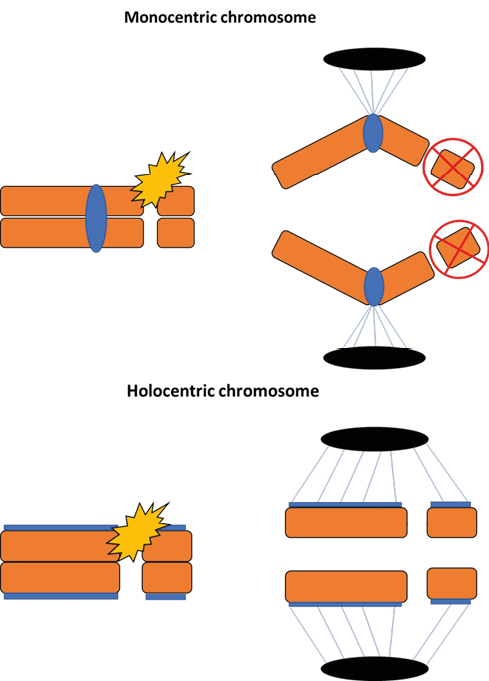
Figure 3 – Consequences of fragmentation in monocentric and holocentric chromosomes. In monocentric chromosomes (above), a double-strand break caused by the presence of a clastogenic agent (e.g.: gamma radiation) results in the formation of acentric fragments (without centromeres), which are normally lost during cell division. In monocentric chromosomes (below), the fragments generated after a double-strand break are recognized by the microtubules of the spindle fibers due to the presence of kinetochores and segregate normally during anaphase.
Several studies have deliberately or indirectly compared the performance of holocentric and monocentric organisms when exposed to clastogenic environments. By subjecting 488 samples representing 82 families of Arthropoda to different levels of ionizing radiation, researchers demonstrated that holocentric chromosomes are less sensitive and monocentric chromosomes are more responsive to high doses of radiation. Furthermore, males and adults are more tolerant to this mutagen when compared to females and juveniles72. Created in the late 1960s and located in northern Wisconsin, United States, Enterprise Radiation Forest allows for field assessments of the effects of gamma radiation on forest ecosystems. When investigating the response of different plant species to a zone with high levels of radiation, researchers detected drastic reductions in biomass production for most plant species studied, with the exception of Luzula acuminata (a recognized holocentric species), which showed an increase in biomass production when kept in the same area73.
The colonization of terrestrial environments by marine eukaryotes required abandonment of the waters and consequent exposure to more intense cosmic radiation. The fact that members of the lineage Zygnematophyceae (a class of green algae ancestral to terrestrial plants) have holocentric chromosomes makes some authors raise the idea that holocentrism was fundamental for the terrestrialization of plants, minimizing the effects of radiation (particularly UV radiation) and desiccation on DNA74. Furthermore, research involving fossil records and molecular data showed that the first independent terrestrialization events in animals involved groups of millipedes, followed by insects, arachnids, roundworms, water bears and onychophores75. As holocentric chromosomes have already been identified in four of these six taxa (see topic Holocentric chromosomes in animals and plants), it is likely that holocentrism also played a key role in the successive occupation of land by animals.
Some authors also hypothesize that the presence of chromosomes with diffuse centromeres constitutes an important defense mechanism for phytophagous insects. When their plant tissue is attacked, some plants are able to produce secondary metabolites that induce numerous DNA damage, including chromosomal fragmentation76. The ability of Mizus persicae nicotianae (popularly known as the tobacco aphid) to form colonies on the leaves of Nicotiana tabacum and to resist nicotine – a volatile alkaloid with a clastogenic effect – is attributed to its holocentric condition77.
The presence of diffuse centromeres along chromosomes per se cannot, however, be considered an adaptive advantage as a rule. The low rates of crossing over characteristic of holocentric chromosomes result in decreased diversity and, consequently, a limited ability to adapt to selective pressures74. In theory, holocentric taxa are expected to have a large variation in chromosome number due to the potential maintenance of the kinetic activity of fragments after chromosomal breaks. A greater number of chromosomes could mean a greater contribution of chromosomal recombinations during anaphase I, which would compensate for the low frequency of crossing over. However, the variation in the number of chromosomes between holocentric and monocentric organisms does not seem to differ drastically. The tolerance of chromosomes with diffuse kinetochores to fragmentation may also mean a suppression of the potential of clastogenic agents for the speciation process. In fact, one study showed the absence of a positive correlation between holocentrism and the rate of diversification or species richness78-79.
The higher rate of recombination in monocentric chromosomes (evidenced by the occurrence of multiple chiasmata per bivalent) results in an increase in genetic variability, thus facilitating the response to selection and, eventually, speciation69. This is the argument usually used to explain the predominance of the monocentric condition among eukaryotes. Furthermore, holocentrism does not seem to be the only way living organisms have been found to deal with environmental instability. The term “extremophile” is used to designate representatives of the Bacteria, Archaea and Eukarya domains that can survive under low or high temperatures, highly acidic or highly basic environments, high pressure, desiccation and/or high salinity80. In this group are the tardigrades, a phylum of apparently monocentric microscopic animals that can tolerate high rates of ionizing radiation due to the presence of the Dsup protein (“Damage suppressor”) in parts of their chromatin81-85. Archaebacteria of the genus Halobacterium are protected from cell damage caused by exposure to gamma radiation thanks to the presence of pigments in their cell membrane, especially bacterioruberin86.
6 FINAL CONSIDERATIONS
The difficulty or lack of interest in studying some organisms cytogenetically (especially those with no eminent economic importance and/or belonging to more basal groups) leads us to believe that the fraction of so-called holocentric species in relation to the totality of eukaryotes is underestimated. Although the observation of the behavior of chromosomes during cell division is historically the most used strategy for the identification of holocentric chromosomes, the amount and distribution of key kinetochore proteins seem to be the best indicators for holocentrism. Much of what we know about holocentric chromosomes in terms of structure and behavior comes from studies involving the model organism Caenorhabditis elegans. However, their presence in phylogenetically distant and unrelated lineages requires caution in establishing generalizations for other eukaryote taxa. The detection of spindle fibers anchored at the ends of chromatids during telokinetic meiosis and in regions that are not equivalent to those occupied by typical proteins of the kinetochore molecular apparatus in the holocentric plant species Cuscuta europaea suggests the existence of possibilities of kinetic activity generated by processes other than the typical binding of kinetochore proteins to microtubules.
Aspects related to the origin and evolution of holocentric chromosomes still remain uncertain, requiring further investigations. Although the adaptive advantages generated by their presence are undeniable (especially in clastogenic environments), and their apparent role in the occupation of the terrestrial environment by animals and plants, the typical behavior of chromosomes with diffuse kinetochores seems to be enough to make them the exception and not the rule in the Eukarya domain.
REFERENCES
- Sybenga, J. Plant Cytogenetics. Heidelberg (Berlin): Springer; 1992.
- Appels R, Morris R, Gill BS, May CE. Chromossome Biology. New York: United States: Springer & Science Business Media; 1998.
- Dernburg AF. Here, there, and everywhere: Kinetochore function on holocentric chromosomes. J Cell Bio. 2001;153(6):33-8.
- Guerra M, Cabral G, Cuacos M, González-García M, González-Sánchez M, Vega J et al. Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet. Genome Res. 2010;129(1-3):82-96.
- Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641-53.
- Schrader F. Notes on the mitotic behavior of long chromosomes. Cytologia. 1935;6(5):422-30.
- Chickering AM. A preliminary study of spermatogenesis of Belastoma (Zaita) fluminea. Trans Am Microsc Soc. 1916;35(1):45-56.
- Melters DP, Paliulis LV, Korf IF, Chan SWL. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579-93.
- Zedek F, Veselý P, Horová L, Bures P. Flow cytometry may allow microscope-independent detection of holocentric chromosomes in plants. Sci. Rep. 2016;6(1):1-8.
- Sheikh SA, Kondo K. Differential staining with orcein, giemsa, CMA and DAPI for comparative chromossome study of 12 species of Australian Drosera (Droseraceae). Am J. Bot. 1995;80(10):1278-86.
- Feitoza L, Costa L, Guerra M. Condensation patterns of prophase/prometaphase chromosome are correlated with H4K5 histone acetylation and genomic DNA contents in plants. PLoS One. 2017;12(8):e0183341.
- Ray JH, Venketeswaran S. Constitutive heterochromatin distribution in monocentric and polycentric chromosomes. Chromosoma. 1978;66(4):341-50.
- Guerra M, García MA. Heterochromatin and rDNA sites distribution in the holocentric chromosomes of Cuscuta approximata Bab. (Corvolvulaceae). Genome. 2004;47(1):134-40.
- Vanzela ALL, Cuadrado A, Guerra M. Localization of 45S rDNA and telomeric sites on holocentric chromosomes of Rhynchospora tenuis Link (Cyperaceae). Genet. Mol. Biol. 2003;26(2):199-201.
- Silva CRM, Quintas CC, Vanzela ALL. Distribution of 45S and 5S rDNA sites in 23 species of Eleocharis (Cyperaceae). Genetica. 2010;138:951-57.
- Nagaki K, Kashihara K, Murata M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell. 2005;17(7):1886-93.
- Braselton JP. The ultrastructure of the non-localized kinetochores of Luzula and Cyperus. Chromosoma, 1971;36(1):89-99.
- Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L. Phosphorylation of histone H3 in plants – A dynamic affair. Biochim. Biophys. Acta Gene Struc. Expre. 2007;1769(5-6):308-15.
- Manzanero S, Arana P, Puertas MJ, Houben A. The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma. 2000;109(5):308-17.
- Guerra M, Brasileiro-Vidal AC, Arana P, Puertas MJ. Mitotic microtubule development and histone H3 phosphorylation in the holocentric chromosomes of Rhynchospora tenuis (Cyperaceae). Genetica. 2006;126:33-41.
- Gernand D, Demidov D, Houben A. The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono – and polycentric chromosomes. Cytogenet. Genome Res. 2003;101(2):172-76.
- Haizel T, Lim YK, Leitch AR, Moore G. Molecular analysis of holocentric centromeres of Luzula species. Cytogenet. Genome Res. 2005;109(1-3):134-43.
- Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14(8):1691-1704.
- Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163(3):1221-25.
- Powers J, Rose DJ, Saunders A, Dunkelbarger S, Strome S, Saxton WM. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J Cell Biol. 2004;166(7):991-1001.
- Stear JH, Roth MB. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 2002;16(12);1498-1508.
- Bures P, Zedek F, Markova M. Holocentric chromosomes. In: Greilhuber J, Dolezel J, Wendel J, editors. Plant Genome Diversity Volume 2. 1st ed. Viena (Austria): Springer; 2013. p. 187-208.
- Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458(7240):852-58.
- Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141(1):159-79.
- Halkka O. Recombination in six homopterous families. Evolution. 1964;18(1):81-8.
- Nokkala S, Kuznetsova VG, Maryanska-Nadachowska A, Nokkala C. Holocentric chromosomes in meiosis I. Restriction of the number of chiasmata in bivalents. Chromosome Res. 2004;12:733-39.
- Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86(3):409-28.
- Nordenskiold H. Tetrad analysis and the course of meiosis in three hybrids of Luzula campestris. Hereditas. 1961;47(2):203-38.
- Dumont J, Oegema K, Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Bio. 2010;12(9):894-901.
- Schvarzstein M, Wignall SM, Villeneuve AM. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 2010;24(3):219-28.
- Ogawa K. Chromosome studies in the Myriapoda V. A chromosomal survey in some chilopods with a cyto-taxonomic consideration. Jpn. J. Genet. 1953;28(1):12-18.
- Rezác M, Král J, Pekár S. The spider genus Dysdera (Araneae, Dysderidae) in Central Europe: revision and natural history. J. Arachnol. 2007;35(3):432-62.
- Benavente R, Wettstein R. Ultrastructural characterization of the sex chromosomes during spermatogenesis of spiders having holocentric chromosomes and a long diffuse stage. Chromosoma. 1980;77(1):69-81.
- Benavente R. Holocentric chromosomes of arachnids: presence of kinetochore plates during meiotic divisions. Genetica. 1982;59(1):23-27.
- Mattos VF, Carvalho LS, Carvalho MA, Schneider MC. Insights into the origin of the high variability of multivalent-meiotic associations in holocentric chromosomes of Tityus (Archaeotityus) scorpions. PLoS One. 2018;13(2):1-23.
- Hill CA, Guerrero FD, Van Zee JP, Geraci NS, Walling JG, Stuart JJ. The position of repetitive DNA sequence in the southern cattle tick genome permits chromosome identification. Chromosome Res. 2009;17:77-89.
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479(7374):487-92.
- Nokkala S, Laukkanen A, Nokkala C. Mitotic and meiotic chromosomes in Somatochlora metallica (Cordulidae, Odonata). The absence of localized centromeres and inverted meiosis. Hereditas. 2002;136(1):7-12.
- Kuznetsova VG, Maryańska-Nadachowska A, Shapoval NA, Anokhin BA, Shapoval AP. Cytogenetic characterization of eight Odonata species originating from the Curonian Spit (the Baltic Sea, Russia) using C-Banding and FISH with 18S rDNA and telomeric (TTAGG) probes. Cytogenet. Genome Res. 2018;153(3);147-57.
- Golub NV, Nokkala S, Kuznetsova VG. Holocentric chromosomes of psocids (Insecta, Psocoptera) analysed by C-banding, silver impregnation and sequence specific fluorochromes CMA 3 and DAPI. Folia Biol. 2004;52(3-4):143-49.
- Murakami A, Imai HT. Cytological evidence for holocentric chromosomes of the silkworms, Bombyx mori and B. mandarina, (Bombycidae, Lepidoptera). Chromosoma. 1974;47(2):167-78.
- Lukhtanov VA, Dinca V, Friberg M, Síchová J, Olofsson M, Vila R et al. Versatility of multivalent orientation, inverted meiosis, and rescued fitness in holocentric chromosomal hybrids. PNAS. 2018;115(41);9610-19.
- Subirana JA, Messeguer X. A satellite explosion in the genome of holocentric nematodes. PLoS One, 2013;8(4):e62221.
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1:15-26.
- Monti V, Manicardi GC, Mandrioli M. Distribution and molecular composition of heterochromatin in the holocentric chromosomes of the aphid Rhopalosiphum padi (Hemiptera: Aphididae). Genetica. 2010;138:077-1084.
- Bardella VB, Pita S, Vanzela ALL, Galvão C, Panzera F. Heterochromatin base pair composition and diversification in holocentric chromosomes of kissing bugs (Hemiptera, Reduviidae). Mem. Inst. Oswaldo Cruz. 2016;111(10):614-24.
- Henikoff S, Thakur J, Kasinathan S, Talbert PB. Remarkable evolutionary plasticity of centromeric chromatin. Cold Spring Harb. Sym. Quant. Biol. 2017;82:71-82.
- Escudero M, Hipp AL, Hansen TF, Voje KL, Luceño M. Selection and inertia in the evolution of holocentric chromosomes in sedges (Carex, Cyperaceae). New Phytol. 2012;195(1)237-47.
- Tanaka N, Tanaka N. Chromosome studies in Chionographis (Liliaceae) I. On the holokinetic nature of chromosomes in Chionographis japonica Maxim. Cytologia. 1977;42(3-4);753-63.
- Hofstatter PG, Thangavel G, Lux T, Neumann P, Vondrak T, Novak P et al. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell. 2022;185(17):3153-3168.
- Marques A, Ribeiro T, Neumann P, Macas J, Novák P, Schubert V et al. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. PNAS. 2015;112(44):13633-13638.
- Ribeiro T, Marques A, Novák P, Schubert V, Vanzela AL, Macas, J et al. Centromeric and non-centromeric satellite DNA organization differs in holocentric Rhynchospora species. Chromosoma. 2017;126:325-335.
- Kynast RG, Joseph JA, Pellicer J, Ramsay MM, Rudall PJ. Chromosome behavior at the base of the angiosperm radiation: karyology of Trithuria submersa (Hydatellaceae, Nymphaeales). Am. J. Bot. 2014;101(9):1447-55.
- Nair RR. Chromosome number analysis in different sex types and open pollinated seedlings of nutmeg (Myristica fragrans Houtt). J. Plant. Crops. 2019;47(3):197-201.
- Schubert V, Neumann P, Marques A, Heckmann S, Macas J, Pedrosa-Harand A, et al. Super-resolution microscopy reveals diversity of plant centromere architecture. Int. J. Mol. Sci. 2020;21(10):1-17.
- Kolodin P, Cempírková H, Bures P, Horová L, Veleba A, Francová J, et al. Holocentric chromosomes may be an apomorphy of Droseraceae. Plant Syst. Evol. 2018;304:1289-96.
- Escudero M, Márquez-Corro JI, Hipp AL. The phylogenetic origins and evolutionary history of holocentric chromosomes. Syst. Bot. 2016;41(3):580-85.
- Mola LM, Papeschi AG. Holokinetic chromosomes at a glance. BAG – J. Bas. Appl. Genet. 2006;17(1):17-33.
- Greilhuber J. Chromosomes of the monocotyledons (general aspects). In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. 1st ed. Surrey (United Kingdom) Kew Royal Botanic Gardens; 1995. p 379-414.
- Villasante A, Abad JP, Méndez-Lago M. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. PNAS. 2007;104(25):10542-47.
- Senaratne AP, Cortes-Silva N, Drinnenberg IA (2022, July). Evolution of holocentric chromosomes: drivers, diversity, and deterrents. Semin Cell Dev Biol. 2022;(127):90-99.
- Cangiano G, Volpe AL. Repetitive DNA sequences located in the terminal portion of the Caenorhabditis elegans chromosomes. Nucleic Acids Res. 1993;21(5):1133-39.
- Neumann P, Navrátilová A, Schroeder-Reiter E, Koblížková A, Steinbauerová V, Chocholová E et al. Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012;8(6):e1002777.
- Márquez-Corro JI, Escudero M, Luceño M. Do holocentric chromosomes represent an evolutionary advantage? A study of paired analyses of diversification rates of lineages with holocentric chromosomes and their monocentric closest relatives. Chromosome Res. 2018;26:139-152.
- Rivi M, Monti V, Mazzoni E, Cassanelli S, Panini M, Bizzaro D et al. Karyotype variations in Italian populations of the peach-potato aphid Myzus persicae (Hemiptera: Aphididae). Bull Entomol Res. 2012;102(6):663-671.
- Mandrioli M, Carlo Manicardi G. Unlocking holocentric chromosomes: new perspectives from comparative and functional genomics? Current Genomics. 2012;13(5):343-349.
- Zedek F, Bures P. Pest arthropods with holocentric chromosomes are more resistant to sterilizing ionizing radiation. Rad. Res. 2019;191(3):255-61.
- Zavitkovski J, Salmonson BJ. Effects of gamma radiation on biomass production of ground vegetation under broadleaved forests of Northern Wisconsin. Rad. Bot. 1975;15(4):337-48.
- Zedek F, Bures P. Holocentric chromosomes: from tolerance to fragmentation to colonization of the land. Ann. Bot. 2018;121(1):9-16.
- Rota-Stabelli O, Daley AC, Pisani D. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr. Biol. 2013;23(5):392-98.
- Mandrioli M, Carlo Manicardi G. Unlocking holocentric chromosomes: new perspectives from comparative and functional genomics? Curr. Genomics. 2012;13(5):343-49.
- Kati AN, Mandrioli M, Skouras PJ, Malloch GL, Voudouris CC, Venturelli M et al. Recent changes in the distribution of carboxylesterase genes and associated chromosomal rearrangements in Greek populations of the tobacco aphid Myzus persicae nicotianae. Biol. J. Linn Soc. 2014;113(2):455-70.
- Márquez-Corro JI, Escudero M, Luceño M. Do holocentric chromosomes represent an evolutionary advantage? A study of paired analyses of diversification rates of lineages with holocentric chromosomes and their monocentric closest relatives. Chromosome Res. 2018;26:139-52.
- Cook LG. Extraordinary and extensive karyotypic variation: a 48-fold range in chromosome number in the gall-inducing scale insect Apiomorpha (Hemiptera: Coccoidea: Eriococcidae). Genome. 2000;43(2):255-63.
- Rothschild LJ, Mancinelli RL. Life in extreme environments. Nature. 2001;409(6823):1092-101.
- Sugiura K, Arakawa K, Matsumoto M. Distribution of Macrobiotus shonaicus Stec, Arakawa & Michalczyk, 2018 (Tardigrada: Eutardigrada: Macrobiotidae) in Japan. Zootaxa. 2020;4767(1):56-70.
- Jeffery NW, Oliveira IS, Gregory TR, Rowell DM, Mayer G. Genome size and chromosome number in velvet worms (Onychophora). Genetica. 2012;140:497-504.
- Rebecchi L, Altiero T, Bertolani R. Banding techniques on tardigrade chromosomes: the karyotype of Macrobiotus richtersi (Eutardigrada, Macrobiotidae). Chromosome Res. 2002;10:437-43.
- Hashimoto T, Horikawa DD, Saito Y, Kuwahara H, Kozuka-Hata H, Shin-I T et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nature Communications. 2016;7(1):1-14.
- Hashimoto T, Kunieda T. DNA protection protein, a novel mechanism of radiation tolerance: lessons from tardigrades. Life. 2017;7(2):1-11.
- Kottemann M, Kish A, Iloanusi C, Bjork S, DiRuggiero J. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles. 2005;9:219-27.
* Master in Genetics and Plant Breeding. “Luiz de Queiroz” College of Agriculture, University of São Paulo, Genetics Department.
† Bachelor of Biotechnology. “Luiz de Queiroz” College of Agriculture, University of São Paulo, Genetics Department,.
‡ Professor, PhD in Evolution Ecology and Population Biology. “Luiz de Queiroz” College of Agriculture, University of São Paulo, Genetics Department.